This tutorial is an extension of the preceding tutorial #4 and makes extensive reference to it; thus it is strongly advisable that the student takes the two in the intended order.
In an experiment aimed at determining the equilibrium constant of the reaction P + X <==> PX, the following data set has been collected:
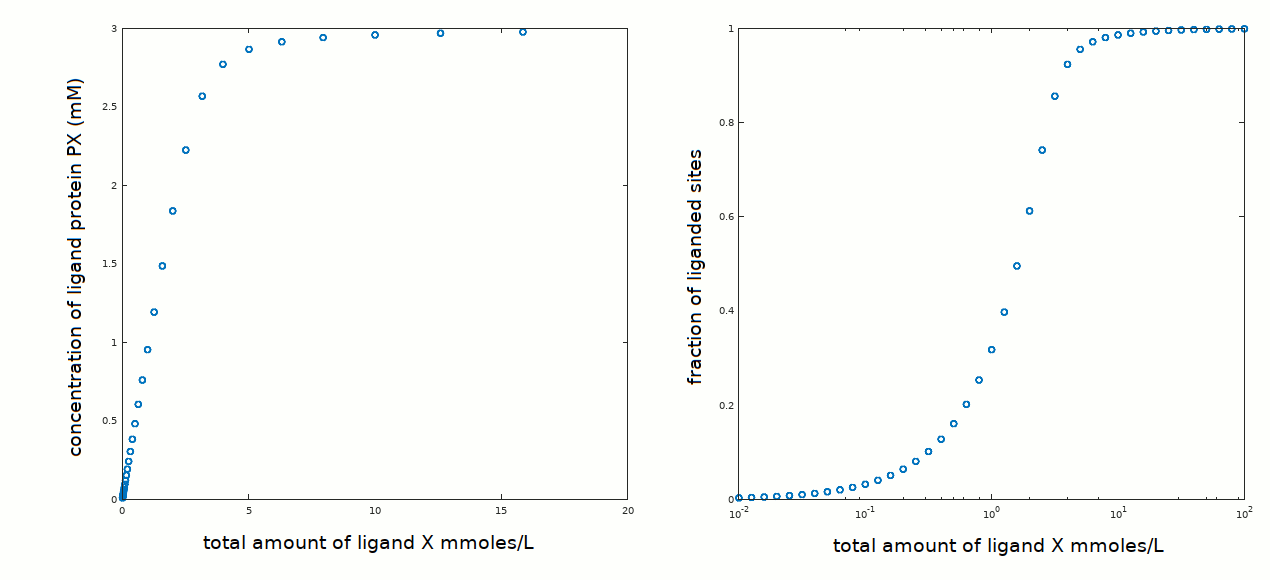
The protein concentration was 3 mM, similar to the ligand concentration required to observe binding. We know from preceding measurements that the protein is pure and monomeric in solution, thus we do not expect cooperativity, positive or negative, or chemical heterogeneity (see tutorials #2 and #3).
How would you proceed with data analysis (click to select)?
1) Wow! These are strange plots indeed! Give me a suggestion
2) Fit eq. Y = [X]/(K+[X]) to the experimental data in order to find the value of K that minimizes the sum of the squared residuals.
3) Linearize the data using Hill's equation log(Y/(1-Y))= n log([X]) - n log(K) then draw the best straight line through the data points in order to find K and n.