answer 1:6 - Correct! The protein presents either chemical heterogeneity or negative cooperativity.
Negative cooperativity, intramolecular heterogeneity and presence of a mixture of isoforms are three possible causes of a broader ligand binding isotherm. These three conditions cannot be distinguished from each other on the basis of equilibrium experiments alone, but require some structural information, e.g. by electrophoresis.
1) Negative cooperativity occurs when the protein is an oligomer made up of identical or at least functionally equivalent subunits, in which binding of the ligand to the first site decreases the ligand affinity of the last site. If we consider as an example the case of a homodimer, the ligand binding reaction can be described by two steps as follows:
If we run an electrophoretic analysis of the native (non-denatured) protein we observe that it migrates as a single band; moreover the MW in solution corresponds to the dimer. If we can separate the monomers they may be identical.
The equation describing ligand saturation for this model is as follows:
2) Intramolecular heterogeneity occurs when the protein is an oligomer of different and non equivalent subunits. In this case each subunit reacts with its own affinity and the one with higher ligand affinity responds to lower ligand concentrations, that with lower affinity to higher concentration. As a consequence the ligand binding isotherm is broader than in the case of a non-cooperative monomeric protein. If the protein is a heterodimer the same equation as in case 1 applies, except that the two binding sites are distinct and the mathematical treatment does not require statistical factors. If we run an electrophoretic analysis of the native (non-denatured) protein we observe that it migrates as a single band; moreover the MW in solution corresponds to the dimer. If we can separate the monomers they result different.
The equation describing ligand saturation for this model, assuming two types of subunits, is as follows:
3) Presence of isoforms. Some proteins are prepared as mixtures of similar but not identical isoforms. All isoforms bind the ligand but they have different affinities and each of them presents its own equilibrium constant. If we run an electrophoretic analysis of the native (non-denatured) protein we observe that it yields two or more bands; the MW in solution corresponds to the monomer. Moreover the quantitative composition of the mixture may vary among different preparations. The equation for this model is identical to that for case 2.
it is important to consider that the 3 cases described above are not mutually exclusive; e.g. one may have a mixture of isoforms forming hetero-oligomers, as in the case of the M and H subunits of tetrameric lactate dehydorgenase.
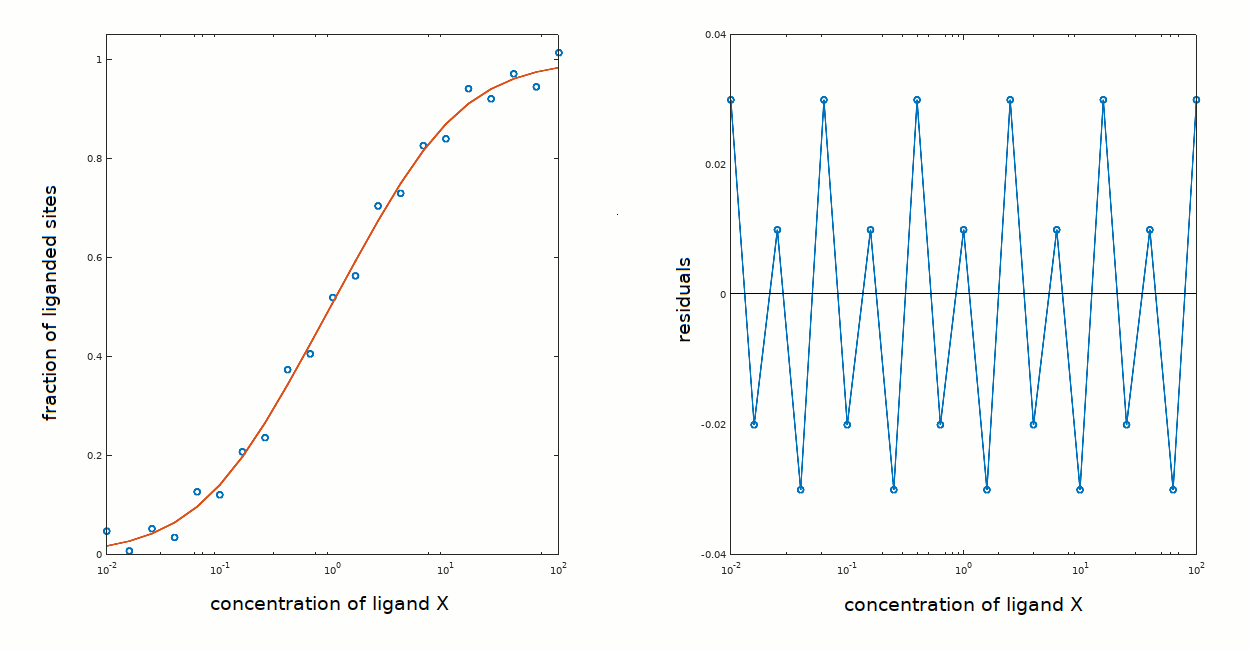
Whatever the answer, the ligand binding isotherm one may calculate for the above cases yields a satisfactory description of the experimental data:
This tutorial is complete. Back to table of content